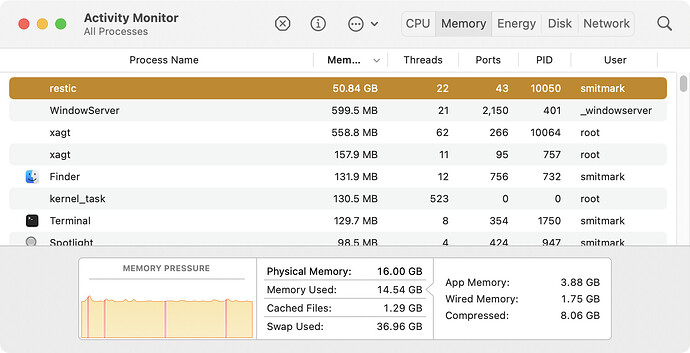
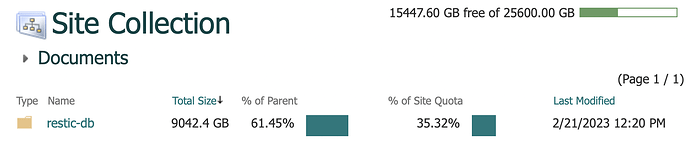After awhile, it continued, and I got a ton of errors like this:
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina122_NovogeneNGS_150PE_M53-138/M55-84_Illumina122_RawDataFiles/M54-89_S0_L001_R2_001.fastq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina122_NovogeneNGS_150PE_M53-138/M55-84_Illumina122_RawDataFiles/M54-89_S0_L001_R2_001.fastq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/M26-124_Illumina80_NovogeneNGS_PE150read60M/raw_data/M26_125_2.fq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/M26-124_Illumina80_NovogeneNGS_PE150read60M/raw_data/M26_125_2.fq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina122_NovogeneNGS_150PE_M53-138/M55-84_Illumina122_RawDataFiles/M54-89_S0_L001_R1_001.fastq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina122_NovogeneNGS_150PE_M53-138/M55-84_Illumina122_RawDataFiles/M54-89_S0_L001_R1_001.fastq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq (1).gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq (1).gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq (1).gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina91_OHSUMPSSR_NextSeq75single_Kazu/LIB190731HN/Undetermined_S0_R1_001.fastq (1).gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina82_NovogeneNGS_PE150read60M/RawData/M27_59_2.fq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina82_NovogeneNGS_PE150read60M/RawData/M27_59_2.fq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina82_NovogeneNGS_PE150read60M/RawData/M27_59_2.fq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina82_NovogeneNGS_PE150read60M/RawData/M27_59_2.fq.gz: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina94_OHSUMPSSR_NextSeq75single/M36-17_Illumina94Sam_PeptideExt_go133_2OUTPUT.txt: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina94_OHSUMPSSR_NextSeq75single/M36-17_Illumina94Sam_PeptideExt_go133_2OUTPUT.txt: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina94_Sam_PeptideExt/M36-17_Illumina94Sam_PeptideExt_go133_2OUTPUT.txt: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina94_Sam_PeptideExt/M36-17_Illumina94Sam_PeptideExt_go133_2OUTPUT.txt: no such file or directory
ignoring error for /Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina84_NovogeneNGS_PE150read250M_Sunghee/M27_93_1.fq.gz: open /Volumes/042122_DL_Backup2/Volumes/Padlock_DT/BoxSyncBackUp_04292022afterUse/Experiments/IlluminaSequencing/Illumina84_NovogeneNGS_PE150read250M_Sunghee/M27_93_1.fq.gz: no such file or directory
![]()
![]()